 |
Lirong Wu (吴立荣) |
News
- [2025.02] Four papers on, AI4Science, have been accepted by ICLR 2025 (1 Spotlight).
- [2024.12] Three papers on, AI4Science, have been accepted by AAAI 2025 (1 Oral).
- [2024.10] Three papers on, AI4Science, have been accepted by NeurIPS 2024.
- [2024.09] One paper on, graph contrastive learning, has been accepted by TNNLS.
- [2024.07] One paper on, graph knowledge distillation, has been accepted by CIKM 2024.
- [2024.05] Five papers on, AI4Science, have been accepted by ICML 2024 (1 Spotlight).
- [2024.03] One paper on, graph knowledge distillation, has been accepted by TKDE.
- [2024.01] Two papers on, AI4Science and graph SSL, have been accepted by ICLR 2024 (1 Spotlight).
- [2023.12] Three papers on, AI4Science, have been accepted by AAAI 2024.
- [2023.09] Four papers on, AI4Science and video prediction, have been accepted by NeurIPS 2023.
- [2023.06] One paper on, graph augmentation, has been accepted by ECML 2023.
- [2023.04] One paper on, graph knowledge distillation, has been accepted by ICML 2023.
- [2023.03] One paper on, graph structure learning, has been accepted by TNNLS.
- [2022.12] One paper on, heterogeneous graph, has been accepted by TNNLS.
- [2022.11] One paper on, graph knowledge distillation, has been accepted by AAAI 2023 (Oral).
- [2022.09] Two papers on, graph augmentation and attack, have been accepted by NeurIPS 2022 (1 Spotlight).
- [2022.07] One paper on, mixup augmentation, has been accepted by ECCV 2022 (Oral).
- [2022.06] One paper on, class-imbalanced classification, has been accepted by ECML 2022 (Oral).
- [2022.02] One paper on, deep clustering, has been accepted by TNNLS.
- [2022.01] One paper on, graph contrastive learning, has been accepted by WWW 2022.
- [2021.11] One paper on, graph self-supervised learning, has been accepted by TKDE.
- [2021.08] One paper on, label denoising, has been accepted by ACM MM 2021 (Oral).
- [2020.09] One paper on, video compression, has been accepted by TCSVT.
- [2020.09] Got my B.E. degree!
Research Interest
Currently, I focus on the following research topics:- Graph Self-supervised Learning
- Heterogeneous/Homophily Graph Learning
- Graph Knowledge Distillation
- AI4Science (Drug Discovery)
Education
- 2020.09-present Ph.D in CAIRI, Westlake University. Supervisor: Prof. Stan Z. Li
- 2016.09-2020.06 B.E. in ISEE, Zhejiang University. Supervisor: Prof. Kejie Huang
Publications
Selected:
|
|
A Simple yet Effective DDG Predictor is An Unsupervised Antibody Optimizer and Explainer |
|
|
Relation-Aware Equivariant Graph Networks for Epitope-Unknown Antibody Design |
|
|
Learning to Predict Mutational Effects of Protein-Protein Interactions by Microenvironment-aware Hierarchical Prompt Learning |
|
|
GeoAB: Towards Realistic Antibody Design and Reliable Affinity Maturation |
|
|
A Teacher-Free Graph Knowledge Distillation Framework with Dual Self-Distillation |
|
|
MAPE-PPI: Towards Effective and Efficient Protein-Protein Interaction Prediction via Microenvironment-Aware Protein Embedding |
|
|
Decoupling Weighing and Selecting for Integrating Multiple Graph Pre-training Tasks |
|
|
PSC-CPI: Multi-Scale Protein Sequence-Structure Contrasting for Efficient and Generalizable Compound-Protein Interaction Prediction |
|
|
Quantifying the Knowledge in GNNs for Reliable Distillation into MLP |
|
|
Extracting Low-/High- Frequency Knowledge from Graph Neural Networks and
Injecting it into MLPs: An Effective GNN-to-MLP Distillation Framework |
|
|
Knowledge Distillation Improves Graph Structure Augmentation for Graph Neural Networks |
|
|
SimGRACE: A Simple Framework for Graph Contrastive Learning without Data Augmentation |
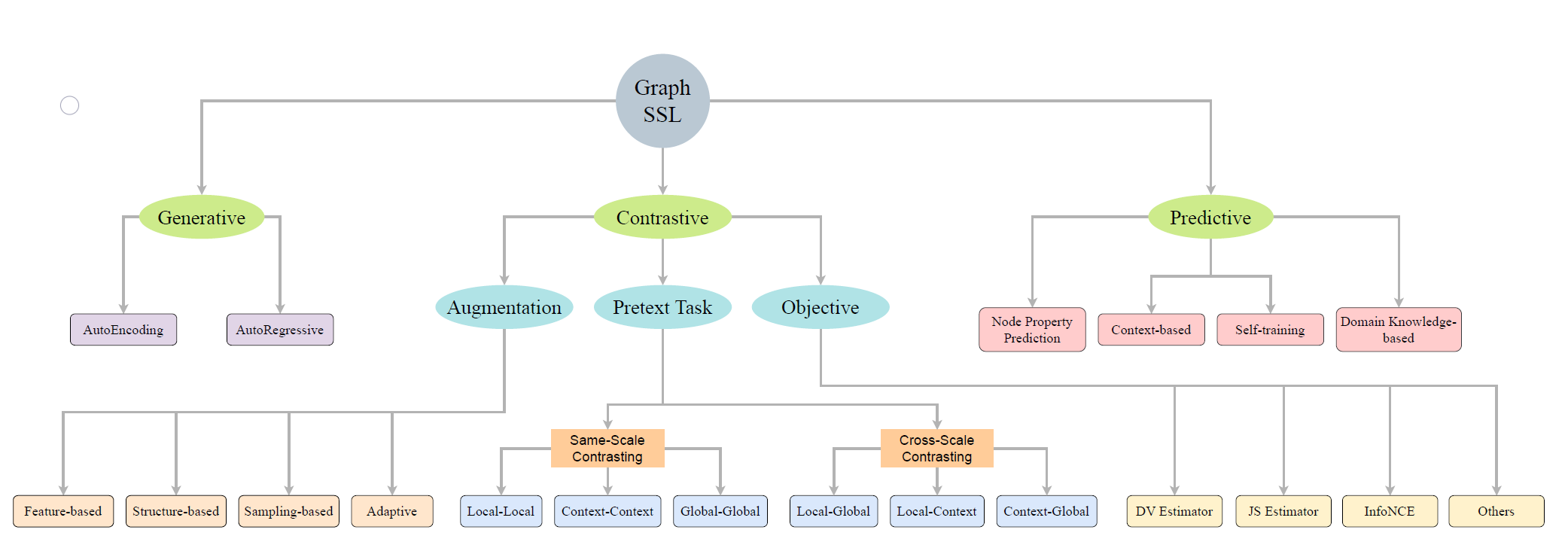 |
Self-supervised Learning on Graphs: Contrastive, Generative,or Predictive |
Publications:
|
|
Teach Harder, Learn Poorer: Rethinking Hard Sample Distillation for GNN-to-MLP Distillation |
|
|
Learning to Augment Graph Structure for both Homophily and Heterophily Graphs |
|
|
GraphMixup: Improving Class-Imbalanced Classification by Self-supervised Context Prediction |
|
|
Learning to Model Graph Structural Information on MLPs via Graph Structure Self-Contrasting |
|
|
Homophily-Enhanced Self-Supervision for Graph Structure Learning: Insights and Directions |
|
|
Beyond Homophily: Relation-Based Frequency Adaptive Graph Neural Networks |
|
|
Deep Clustering and Visualization for End-to-End High-Dimensional Data Analysis |
|
|
Foreground-background Parallel Compression with Residual Encoding for Surveillance Video |
|
|
Re-Dock: Towards Flexible and Realistic Molecular Docking with Diffusion Bridge |
|
|
Towards Reasonable Budget Allocation in Untargeted Graph Structure Attacks via Gradient Debias |
|
|
AutoMix: Unveiling the Power of Mixup for Stronger Classifiers |
|
|
Co-learning: Learning from Noisy Labels with Self-supervision |
Services
Membership:
- IEEE, Student Member, 2019-present
Program committee member | Reviewer
- IEEE Conference on Computer Vision and Pattern Recognition (CVPR)
- International Conference on Computer Vision (ICCV)
- ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD)
- Conference and Workshop on Neural Information Processing Systems (NeurIPS)
- International Conference on Machine Learning (ICML)
- IEEE Transactions on Neural Networks and Learning Systems (TNNLS)